
What is ODTlearn?#
Decision trees are interpretable machine learning models that are routinely used in applications involving classification and prescriptive problems. A decision tree takes the form of a binary tree and at each branching node of the tree, a binary test is performed on a specific feature. Two branches emanate from each branching node, with each branch representing the outcome of the test. If a data point passes (resp. fails) the test, it is directed to the left (resp. right) branch. A label (prediction) or treatment (prescription) is assigned to all leaf nodes for classification and prescriptive problems, respectively. Thus, each path from root to leaf represents a classification or treatment assignment rule that assigns a unique label or treatment to all data points that reach that leaf. The goal in the design of optimal decision trees is to select the best tests to perform at each branching node and which label or treatment to assign to each leaf to maximize the relevant objective.
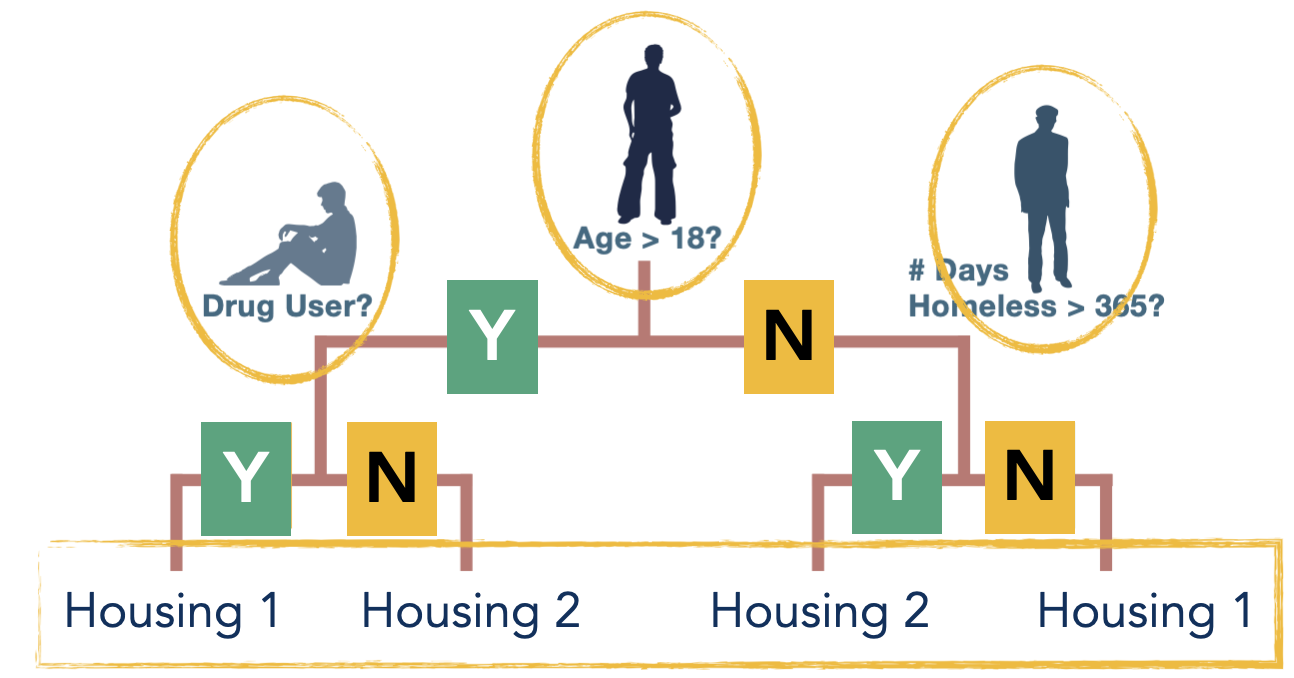
ODTlearn is a Python package for learning various types of decision trees, which is developed based on research out of University of Southern California. ODTlearn uses Mixed-Integer-Optimization (MIO) technology for modeling a variety of decision trees including:
StrongTrees for learning optimal classification trees (Aghaei et al. (2021))
FairTrees for learning optimal classification trees that can incorporate various notions of fairness such as statistical parity, conditional statistical parity, predictive equality, equal opportunity and equalized odds (Jo et al. (2021))
RobustTrees for learning optimal classification trees that are robust against distribution shifts (Justin et al. (2021))
PrescriptiveTrees for learning optimal prescriptive trees (Jo et al. (2021))
Resources for Getting Started#
There are a few ways to get started with ODTlearn:
Read the Installation Guide.
Read the User Guide for an overview of each of the algorithms in this package.
Review the Example Notebooks for each of the methods implemented in ODTlearn.
Documentation Structure#
We provide an overview of the structure of our documentation to help you know where to look when you run into any issues:
Example notebooks walk through fitting decision trees for several toy problems using ODTlearn. Start here if you are new to ODTlearn, or you have a particular type of problem you want to model.
The API Reference contains a complete list of the classes and methods you can use in ODTlearn. Go here to know how to use a particular classifier and its corresponding methods.
The Developer Docs section contains information for people interested in contributing to ODTlearn development or writing an ODTlearn extension. Don’t worry about this section if you are using ODTlearn to solve problems as a user.
Usage Example#
The following script demonstrates how to use the ODTlearn package to fit a StrongTree. For examples of how to use other types of trees please consult the example notebooks and API documentation.
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from odtlearn.FlowOCT import FlowOCT
data = pd.read_csv("./data/balance-scale_enc.csv")
y = data.pop("target")
X_train, X_test, y_train, y_test = train_test_split(
data, y, test_size=0.33, random_state=42
)
stcl = FlowOCT(
depth=1,
time_limit=100,
_lambda=0,
num_threads=None,
obj_mode="acc",
)
stcl.fit(X_train, y_train, verbose=True)
stcl.print_tree()
test_pred = stcl.predict(X_test)
print(
"The out-of-sample acc is {}".format(np.sum(test_pred == y_test) / y_test.shape[0])
)
References#
Aghaei, S., Gómez, A., & Vayanos, P. (2021). Strong optimal classification trees. arXiv preprint arXiv:2103.15965. [arxiv]
Jo, N., Aghaei, S., Benson, J., Gómez, A., & Vayanos, P. (2022). Learning optimal fair classification trees. arXiv preprint arXiv:2201.09932. [arxiv]
Justin, N., Aghaei, S., Gomez, A., & Vayanos, P. (2021). Optimal Robust Classification Trees. In The AAAI-22 Workshop on Adversarial Machine Learning and Beyond. [link]
Jo, N., Aghaei, S., Gómez, A., & Vayanos, P. (2021). Learning optimal prescriptive trees from observational data. arXiv preprint arXiv:2108.13628. [arxiv]